Identify peak tables with multiple databases
Xiaotao Shen PhD (https://www.shenxt.info/)
Created on 2020-03-28 and updated on 2022-09-19
Source:vignettes/multiple_databases.Rmd
multiple_databases.RmdSome time we have multiple databases, so we can use
identify_metabolite_all() functions to identify metabolites
using multiple database.
MS1 data preparation
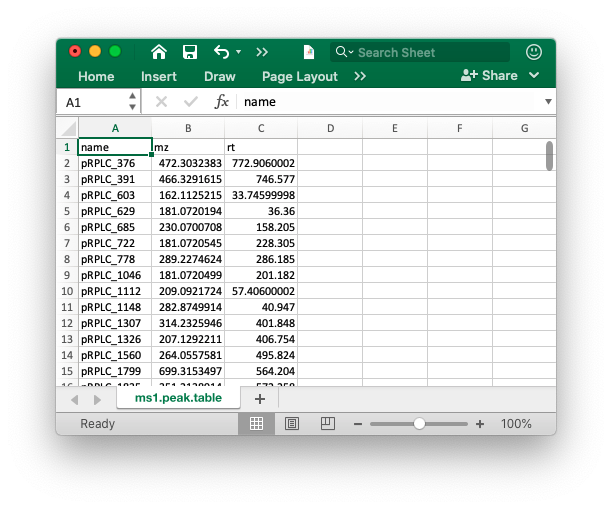
The peak table must contain “name” (peak name), “mz” (mass to charge ratio) and “rt” (retention time, unit is second). It can be from any data processing software (XCMS, MS-DIAL and so on).
MS2 data preparation
The raw MS2 data from DDA or DIA should be transfered to msp, mgf or mzXML format files using ProteoWizard software.
Database
The database must be generated using
construct_database() function. Here we use the databases in
metid packages.
snyder_database_rplc0.0.3: A in-house database, it contains m/z, RT and MS2 spectra information.orbitrap_database0.0.3: A public database fromMassBank. It contains m/z and MS2 spectra information.hmdb_ms1_database0.0.3: A public database fromHMDB, only contains m/z information.
Data organization
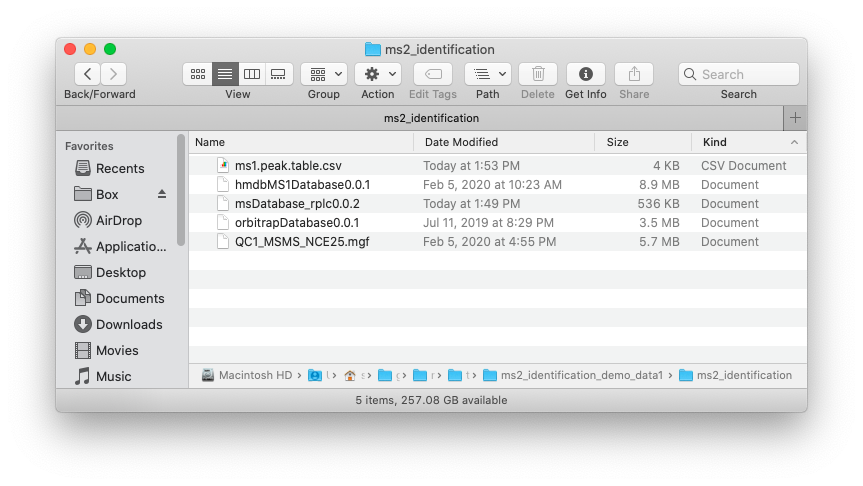
Place the MS1 peak table, MS2 data and databases which you want to use in one folder like below figure shows:
Load demo data
First we load the MS1 peak, MS2 data and databases from
metid package and then put them in a example
folder.
##creat a folder nameed as example
path <- file.path(".", "example")
dir.create(path = path, showWarnings = FALSE)
##get MS1 peak table from metid
ms1_peak <- system.file("ms1_peak", package = "metid")
file.copy(
from = file.path(ms1_peak, "ms1.peak.table.csv"),
to = path,
overwrite = TRUE,
recursive = TRUE
)
##get MS2 data from metid
ms2_data <- system.file("ms2_data", package = "metid")
file.copy(
from = file.path(ms2_data, "QC1_MSMS_NCE25.mgf"),
to = path,
overwrite = TRUE,
recursive = TRUE
)
##get databases from metid
data("snyder_database_rplc0.0.3")
data("orbitrap_database0.0.3")
save(snyder_database_rplc0.0.3, file = file.path(path, "snyder_database_rplc0.0.3"))
save(orbitrap_database0.0.3, file = file.path(path, "orbitrap_database0.0.3"))Now in your ./example, there are files files, namely
ms1.peak.table.csv, QC1_MSMS_NCE25.mgf and
three databases.
Set parameter list
We need to use identify_metabolites_params() functions
to set parameter list for each database.
param1 <-
identify_metabolites_params(
ms1.match.ppm = 15,
rt.match.tol = 15,
polarity = "positive",
ce = "all",
column = "rp",
total.score.tol = 0.5,
candidate.num = 3,
threads = 3,
database = "snyder_database_rplc0.0.3"
)
param2 <- identify_metabolites_params(
ms1.match.ppm = 15,
rt.match.tol = 15,
polarity = "positive",
ce = "all",
column = "rp",
total.score.tol = 0.5,
candidate.num = 3,
threads = 3,
database = "orbitrap_database0.0.3"
)
param3 <- identify_metabolites_params(
ms1.match.ppm = 15,
rt.match.tol = 15,
polarity = "positive",
ce = "all",
column = "rp",
total.score.tol = 0.5,
candidate.num = 3,
threads = 3,
database = "hmdb_ms1_database0.0.3"
)Note: You can set different parametes for each database.
Metabolite identification
All the parameters for three databases should be provided to
parameter.list.
result <- identify_metabolite_all(
ms1.data = "ms1.peak.table.csv",
ms2.data = "QC1_MSMS_NCE25.mgf",
parameter.list = c(param1, param2, param3),
path = path
)Note:
resultis a list, and each element is ametIdentifyClassobject. So you can use the functions formetIdentifylassobject to process it.
Integrate annotation results from different databases
After we get the annotation result list, we then can integrate the annotation results from different databases.
For snyder_database_rplc0.0.3, the annotaiton results
are Level 1 according to MSI.
result[[1]]Then get the annotation table.
annotation_table1 <-
get_identification_table(result[[1]], type = "new", candidate.num = 1)
annotation_table1 %>%
head()For orbitrap_database0.0.3, the annotaiton results are
Level 2 according to MSI.
result[[2]]Then get the annotation table.
annotation_table2 <-
get_identification_table(result[[2]], type = "new", candidate.num = 1)
annotation_table2 %>%
head()For hmdb_ms1_database0.0.3, the annotaiton results are
Level 3 according to MSI.
result[[3]]Then get the annotation table.
annotation_table3 <-
get_identification_table(result[[3]], type = "new", candidate.num = 1)
annotation_table3 %>%
head()Then we should combine them together:
annotation_table1 <-
annotation_table1 %>%
dplyr::filter(!is.na(Compound.name))
dim(annotation_table1)
annotation_table1 <-
data.frame(annotation_table1,
Level = 1,
stringsAsFactors = FALSE)
annotation_table2 <-
annotation_table2 %>%
dplyr::filter(!is.na(Compound.name))
dim(annotation_table2)
annotation_table2 <-
data.frame(annotation_table2,
Level = 2,
stringsAsFactors = FALSE)
annotation_table3 <-
annotation_table3 %>%
dplyr::filter(!is.na(Compound.name))
dim(annotation_table3)
annotation_table3 <-
data.frame(annotation_table3,
Level = 3,
stringsAsFactors = FALSE)If one peak have annotation from three different database, we only contains the annotations with higher confidence.
annotation_table2 <-
annotation_table2 %>%
dplyr::filter(!(name %in% annotation_table1$name))
annotation_table <-
rbind(annotation_table1,
annotation_table2)
annotation_table3 <-
annotation_table3 %>%
dplyr::filter(!(name %in% annotation_table$name))
annotation_table <-
rbind(annotation_table,
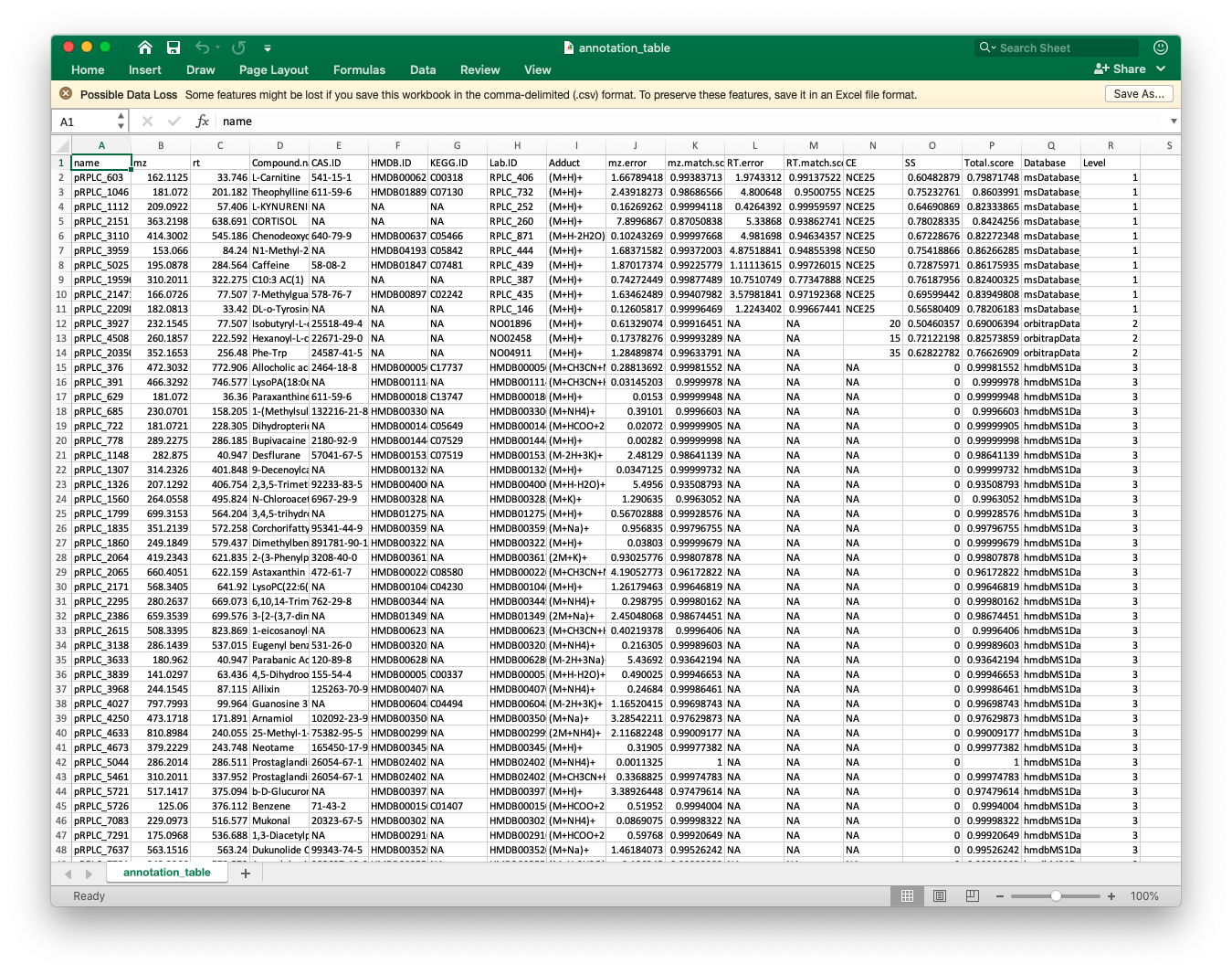
annotation_table3)The annotation_table is the final annotation table.
Then we can output it as csv file.
Session information
sessionInfo()
#> R version 4.2.1 (2022-06-23)
#> Platform: x86_64-apple-darwin17.0 (64-bit)
#> Running under: macOS Big Sur ... 10.16
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> loaded via a namespace (and not attached):
#> [1] rprojroot_2.0.3 digest_0.6.29 R6_2.5.1 jsonlite_1.8.0
#> [5] magrittr_2.0.3 evaluate_0.15 stringi_1.7.8 rlang_1.0.5
#> [9] cachem_1.0.6 cli_3.3.0 rstudioapi_0.14 fs_1.5.2
#> [13] jquerylib_0.1.4 bslib_0.3.1 ragg_1.2.2 rmarkdown_2.14
#> [17] pkgdown_2.0.6 textshaping_0.3.6 desc_1.4.1 tools_4.2.1
#> [21] stringr_1.4.1 purrr_0.3.4 yaml_2.3.5 xfun_0.31
#> [25] fastmap_1.1.0 compiler_4.2.1 systemfonts_1.0.4 memoise_2.0.1
#> [29] htmltools_0.5.2 knitr_1.39 sass_0.4.1