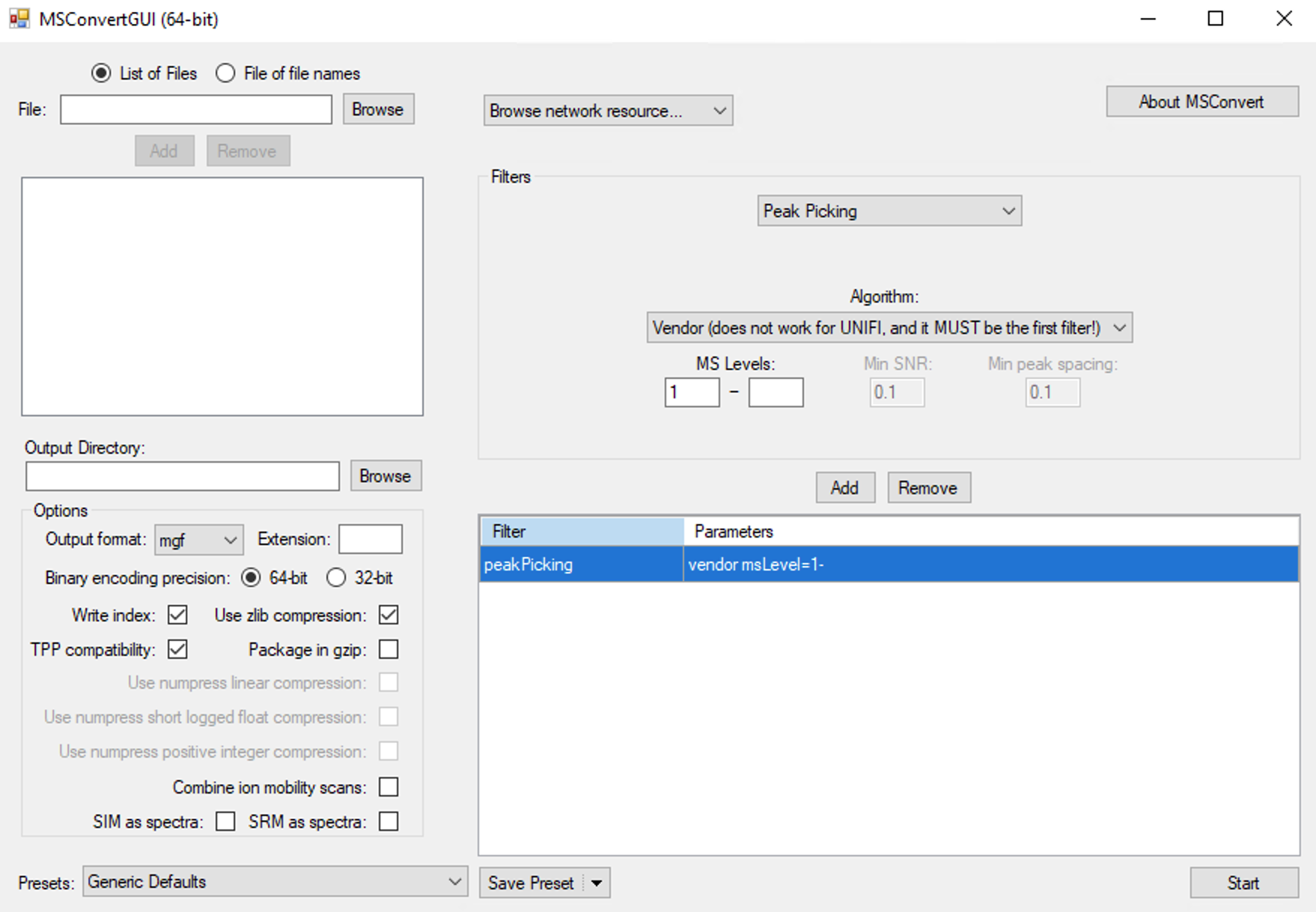
Convert raw data to mzXML/mgf
Xiaotao Shen (https://www.shenxt.info/)
Created on 2021-12-04 and updated on 2022-03-21
data_convert.RmdData preparation
Please place the raw data in one folder according to MS1 and MS2. Then you can convert them using Proteowizard or massconverter package.
Convert data using massconverter
massconverter is a package in tidymass project which can be used to convert mass spectrometry raw data based on docker image of pwid. See more information here.
More inforamtion can be found here.
Session information
sessionInfo()
#> R Under development (unstable) (2022-01-11 r81473)
#> Platform: x86_64-apple-darwin17.0 (64-bit)
#> Running under: macOS Big Sur/Monterey 10.16
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> loaded via a namespace (and not attached):
#> [1] rprojroot_2.0.2 digest_0.6.29 crayon_1.4.2 R6_2.5.1
#> [5] jsonlite_1.7.2 magrittr_2.0.1 evaluate_0.14 stringi_1.7.6
#> [9] rlang_0.4.12 cachem_1.0.6 fs_1.5.2 jquerylib_0.1.4
#> [13] bslib_0.3.1 ragg_1.2.1 rmarkdown_2.11 pkgdown_2.0.1
#> [17] textshaping_0.3.6 desc_1.4.0 tools_4.2.0 stringr_1.4.0
#> [21] purrr_0.3.4 yaml_2.2.1 xfun_0.29 fastmap_1.1.0
#> [25] compiler_4.2.0 systemfonts_1.0.3 memoise_2.0.1 htmltools_0.5.2
#> [29] knitr_1.37 sass_0.4.0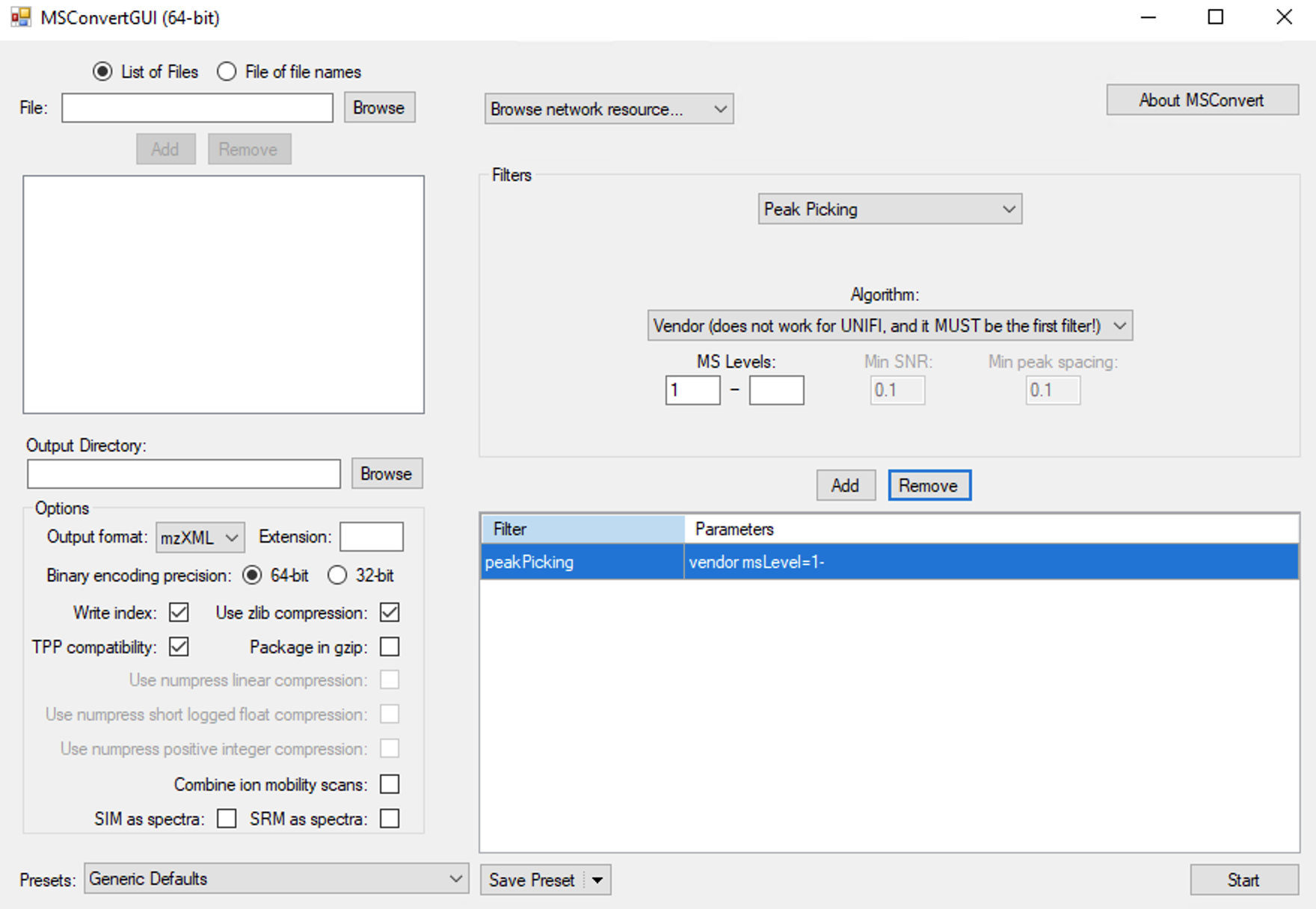 ## Convert MS2 data to
## Convert MS2 data to